De Novo Drug Design using A.I.
We propose a novel generative deep-learning approach for drug design. The model consists of an Encoder-Decoder architecture that translates the inputs into the latent space and a Generator that produces the 3D structure of the system. We propose a stacked Generator architecture that takes the first output and calculates regions of interest. We use the regions of interest to re-sample and generate a second output that is concatenated to the first to produce the prediction of the 3D structure of the molecular system.
An outbreak of the novel coronavirus SARS-CoV-2 has infected millions of people, killed over half million and caused worldwide social and economic disruption.
There are currently no antiviral drugs with clinically proven efficiency nor are there vaccines for its prevention.
The time and effort to create and market a drug or vaccine can span over decades and millions of investment. It is estimated that the drug discovery and development process takes around 10–14 years and more than 1 billion dollars capital in total.
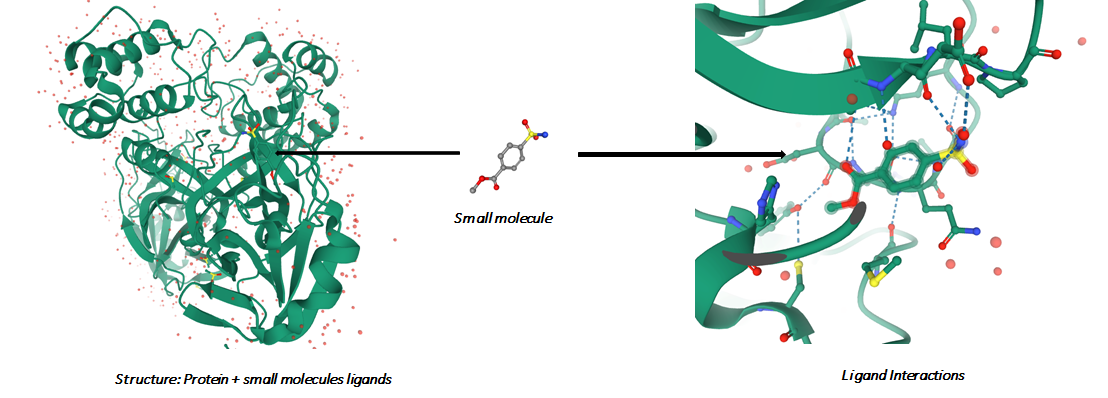
We need to find or build a molecular structure that is able to attach itself to a target protein. It needs to fit within the 3D structure of the target and it needs to produce the correct chemical reactions with the target. The quality of the binding impacts side effects and effectiveness of the treatment
The computer-assisted de novo design of chemical structures offers a viable strategy to reduce efforts and obtain bioactive small molecules. The current computational de novo design methods for generating small molecules suffer from several limitations.
Artificial Intelligence combined with computational power can enable the production of chemically correct structures with a planned biological activity.
In this paper we propose the generation of synthetic molecule structures that optimize the binding affinity to a target (ASYNT-GAN).
We achieve this by leveraging on important milestones in Deep Learning:
•Attention
•Deep Learning on Graphs
•Generative Adversarial Neural Networks
By adopting this approach, we propose a novel way of searching for existing compounds that are suitable candidates.
Similar to question and answer in Natural Language Processing (NLP) we are able to find drugs with highest relevance to a target. We are able to identify substructures that are the most suitable for binding.
Method
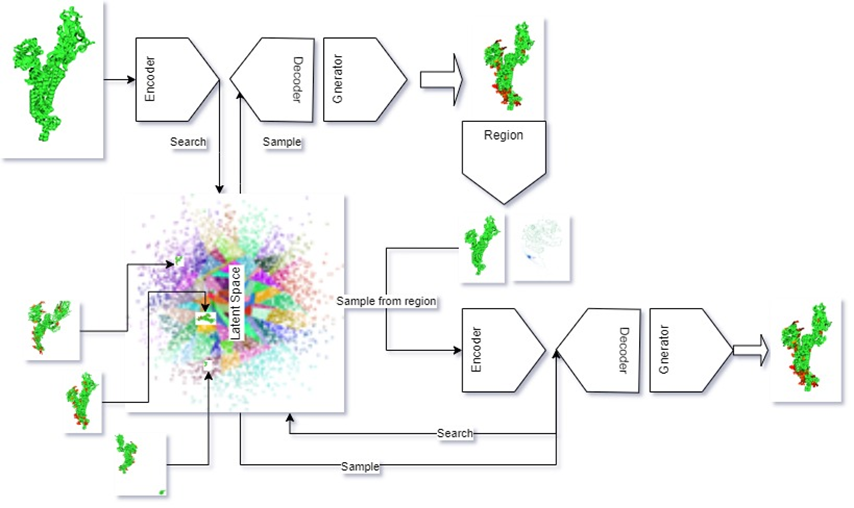
The model consists of an Encoder-Decoder architecture that translates the inputs into the latent space and a Generator that produces the 3D structure of the system. We propose a stacked Generator architecture that takes the first output and calculates regions of interest. We use the regions of interest to re-sample and generate a second output that is concatenated to the first to produce the prediction of the 3D structure of the molecular system.
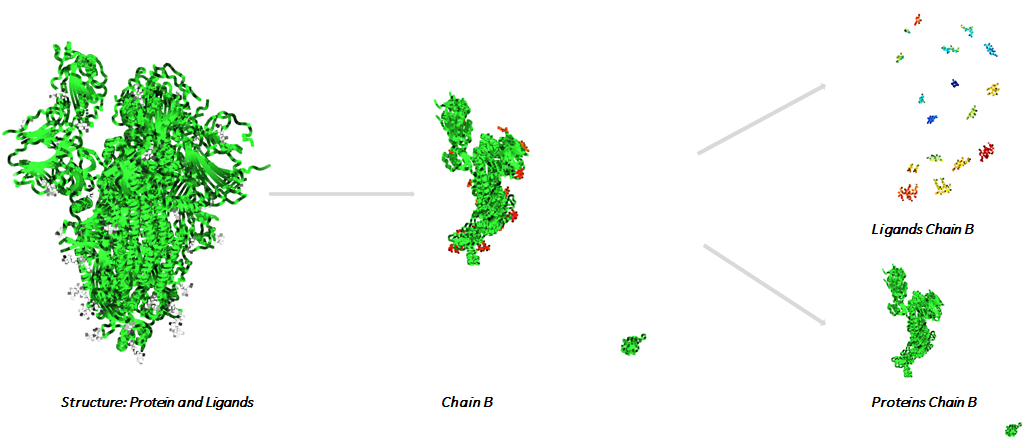
We used a series of viral Proteins of the SARS-CoV-2 from data bank RCSB. The systems that we consider as valid contain ligands that are referenced as Chemical Component with a Drug Bank ID. Each system is split into chains. Each chain is split into proteins and ligands.
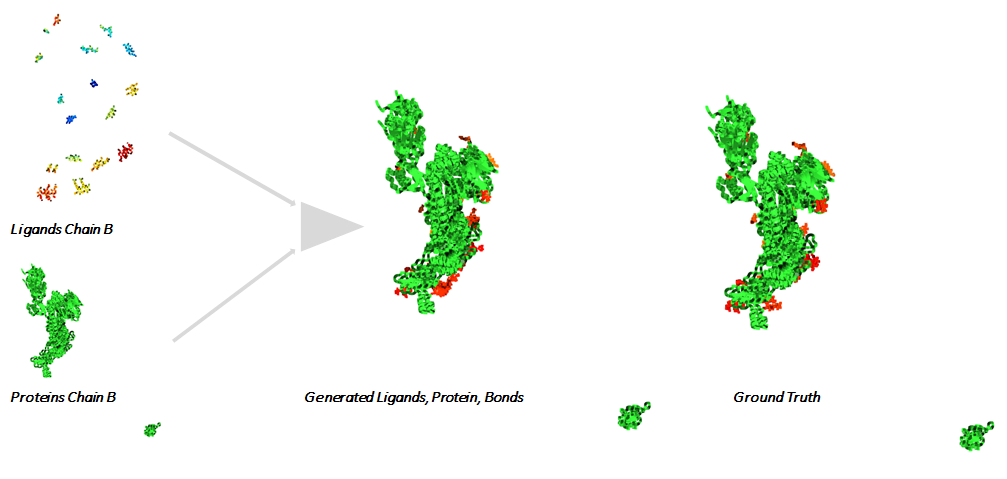
During training we use proteins as input 1 and a sample of the Gaussian distribution as input 2. We have experimented with approaches where Input 2 is a limited number of points sampled from the ligand.
The method generates a full system in 3D space comprising of Ligands, Protein and Ligand Bonds (Ligand Interactions)
Similarity Search
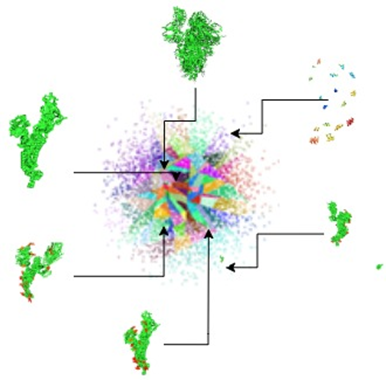
With our method we effectively translate the inputs into the latent space. We can use these properties to index full systems or part of systems and perform a search for similar systems. The embeddings of all the systems are inserted into an index and searched for similarities using Approximate nearest neighbor.
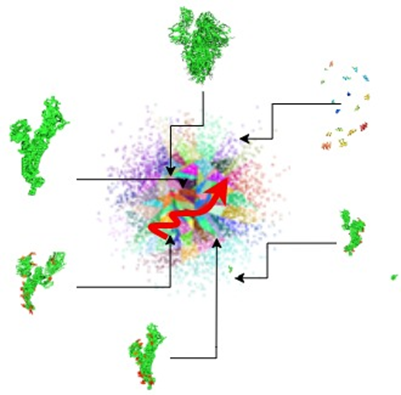
The latent space has structure that can be explored, such as by interpolating between points and performing vector arithmetic between points. For instance we can use the best match from the approximate nearest neighbor search as starting point for a walk through the latent space.
Results
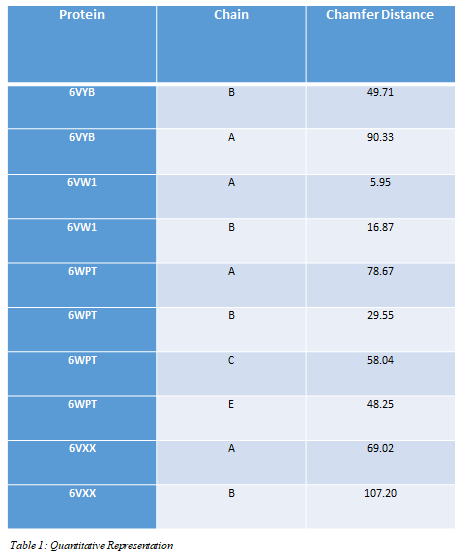
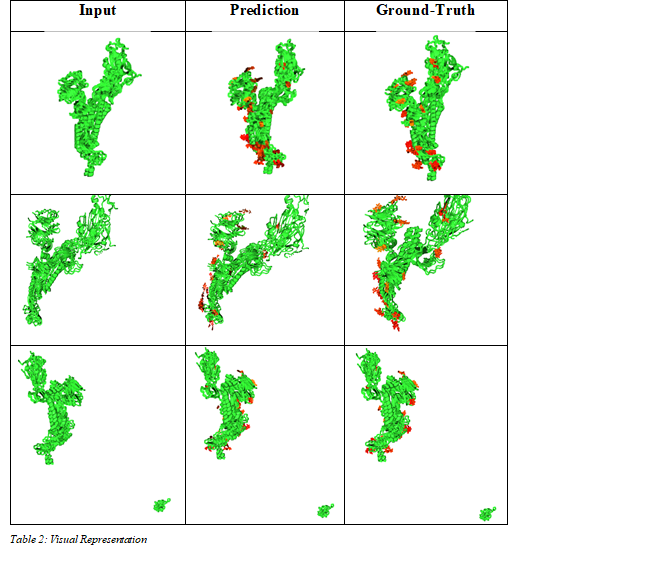
We evaluated our method on a series of viral Proteins of the SARS-CoV-2. We compare quantitative and qualitative performance in Table 1 and Table 2. Quantitative the difference between the generated systems and the ground truth is small. Qualitative our solution achieves good generation of complete structures. The learned representation does generalize to systems beyond the ones used during training.
Conclusions and next steps
Our experiments show that we are able generate complete systems and to generalize to structures of unseen systems.
Translating the input systems into the latent space permits searchability for similar structures and sampling from the latent space for generation.
Topics for future work include integrating the search capabilities in the training process, exploring alternatives for sampling and generating from regions of interest.
